blender-tissue-cartography
What this tool does
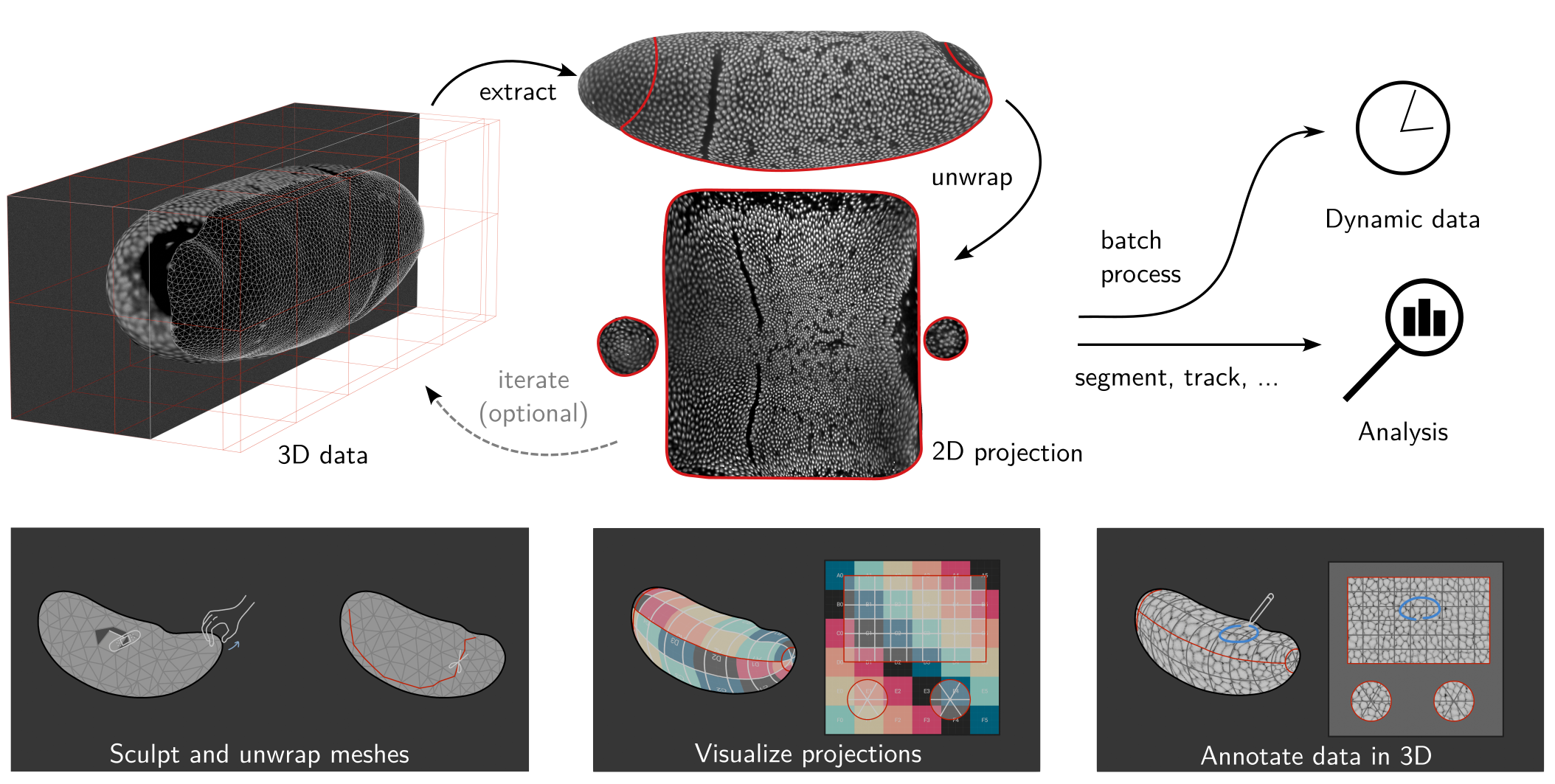
Tissue cartography extracts and cartographically projects curved surfaces from volumetric image data. This turns your 3D data into 2D data, which is much easier to visualize, analyze, and computationally process. Tissue cartography is particularly useful in developmental biology, analyzing 3d microscopy data by taking advantage of many biological tissues’ laminar, sheet-like organization. For more on tissue cartography, see our methods paper, Heemskerk & Streichan 2015 and Mitchell & Cislo 2023.
blender_tissue_cartography comprises an add-on for tissue cartography using the popular 3d creation software blender, a Python library for creating custom/automatized analysis pipelines, and a set of template analysis pipelines/tutorials.
Click here for a 2-minute video demo
Work in progress!
This project is a work in progress and may change rapidly. Please update regularly.
Installation
blender_tissue_cartography comprises both a Blender add-on and a python library.
The add-on does tissue cartography 100% within the Blender graphical user interface. Use the add-on if you quickly want to process a new dataset, or if you are not a coding expert.
The python library allows expert users to develop custom or automatized pipelines.
System requirements Both the Python library and the add-on have no minimum system requirements and can run on any modern laptop. For processing volumetric image data, you will need sufficient RAM (at least ~4x of the size of the volumetric data, e.g. 8GB RAM for a 2GB volumetric .tif file). Blender will run much faster if your computer has a GPU. The MeshLab library which is required for some (non-essential) operations is not available for new ARM Apple computers.
Blender add-on
Install the non-python programs: Blender 4.2 or later (pre-4.2 version will not work) and Ilastik. You do not need to install Python or any Python libraries.
From GitHub, download the file
blender_addon/blender_tissue_cartography-1.0.0-[XXX].zipwhere[XXX]is your operating system (e.g.linux_x64). You do not need to unpack the.zip.- If your operating system is not available, you can also download
blender_addon/blender_tissue_cartography.py. In this case you will need to install the python libraryscikit-imagein Blender’s Python interface.
- If your operating system is not available, you can also download
Install the add-on: Click “Edit -> Preferences -> Add-ons -> Add-on Settings -> Install from disk” and select the
.zipfile you just downloaded (you don’t need to unpack it).Restart Blender. The add-on can now be found under “Scene -> Tissue Cartography”.
Python library
Install the non-python programs: Fiji (optional), Ilastik, Meshlab (optional), and Blender.
Install Python via anaconda/miniconda, if you haven’t already.
Install
blender_tissue_cartography: runpip install blender-tissue-cartographyin a command window.(Optional) Install extra Python library for
pymeshlab, required for some advanced (re)meshing functionality. This package is not available on new ARM Apple computers.- run
pip install pymeshlabin a command window
- run
The project is hosted on pip, with source code on GitHub.
Developer installation
If you want to extend blender_tissue_cartography:
Clone the github repository.
Create a
condaenvironment with all Python dependencies and install theblender_tissue_cartographymodule. Open a command window in theblender-tissue-cartographydirectory and type:conda env create -n blender_tissue_cartography -f environment.ymlconda activate blender_tissue_cartographypip install -e .
(Optional) Install extra Python library for
pymeshlab, required for certain advanced functionality (remeshing and surface reconstruction from within Python).pip install pymeshlab- Note that this package is not available on new ARM Apple computers.
Install nbdev
Known issues
When using the add-on, please save your
.blendproject to disk before using any of the add-on functionality. Otherwise, Blender may crash.3D
.tiffiles loaded into Blender are not saved to the.blend(which would explode in size). If you close Blender, you need to reimport your 3D image data.Please report any issues or bugs you find on GitHub!
Documentation
Datasets and interactive Jupyter notebooks for the tutorials can be downloaded from GitHub
The methods paper explains the general idea of tissue cartography, the design of
blender_tissue_cartography, and shows several examples.
Basic usage
For a complete set of tutorials, see the documentation website.
Tissue cartography workflow
Tissue cartography starts with a 3D, volumetric image.
Create a segmentation of your 3D data to identify the surface you want to extract
Convert the segmentation into a mesh of your surface of interest
Cartographically unwrap the mesh into a 2D plane
Project your 3D data onto the unwrapped mesh
Visualize the results in 3D using Blender or use the 2D projected data for quantitative analysis.
Batch process multiple 3D images (e.g. frames of a movie)
Blender add-on
The Blender add-on allows you to carry out steps 2-5 entirely within Blender. Here is a screenshot using the example Drosophila dataset:
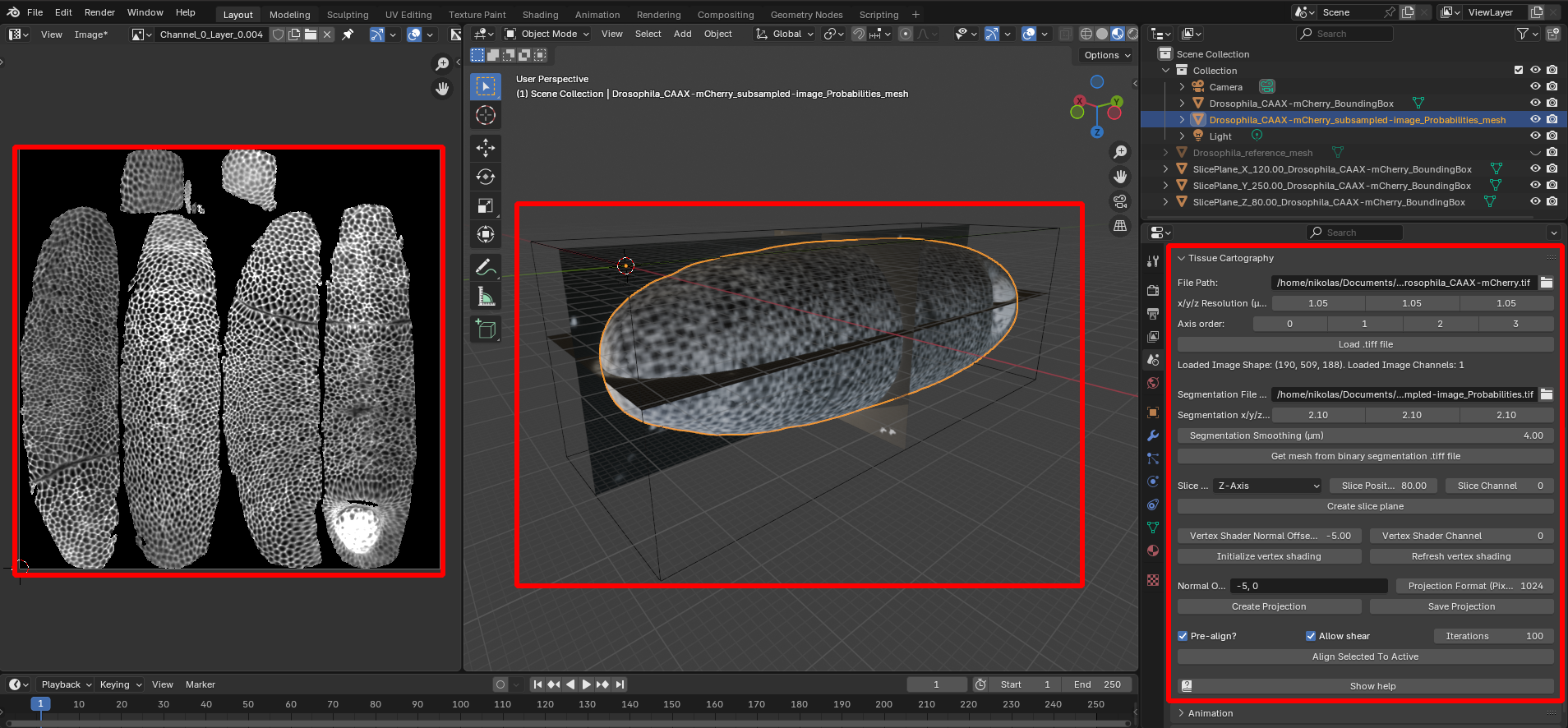
Left: Projected 2D image. Center: 3D view of image data (volume bounding box, image slices, and extracted surface). Right: Tissue Cartography add-on panel.
In Blender, you can edit meshes and cartographic projections interactively - you can create a preliminary projection of your data automatically, and use it as guidance when editing your cartographic map in blender. Here, we edit the “seam” of our cartographic map based on the region occupied by cells during zebrafish epiboly (tutorial 6).
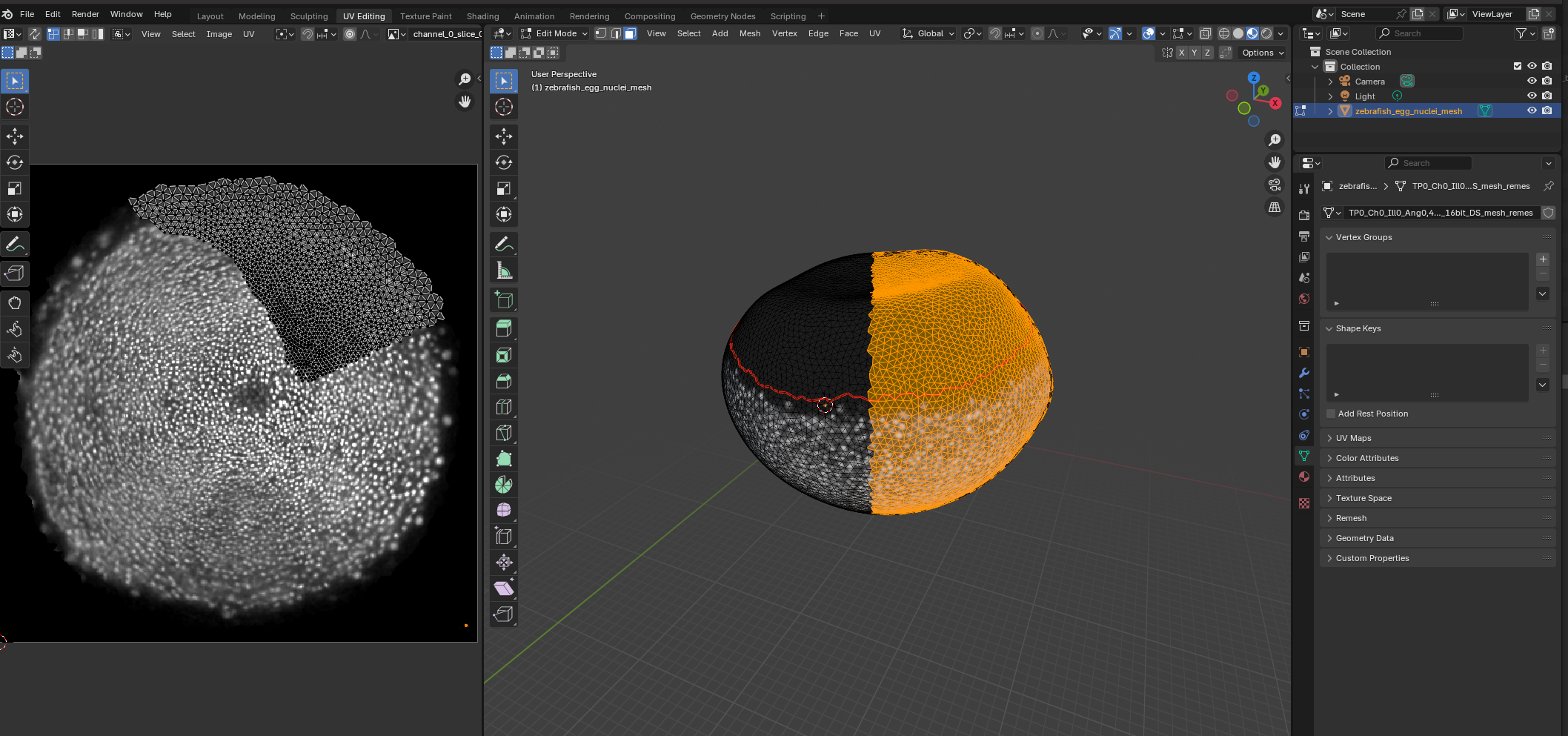
Dynamic datasets
blender_tissue_cartography also allows the creation of cartographic projections of dynamic datasets (i.e. movies), where the surface of interest can move or deform over time. The user creates a cartographic projection for a reference time point which is transferred to all other time points using surface-to-surface registration algorithms. This generates consistent projections across all time points - see tutorials 8 and 9.
Python library
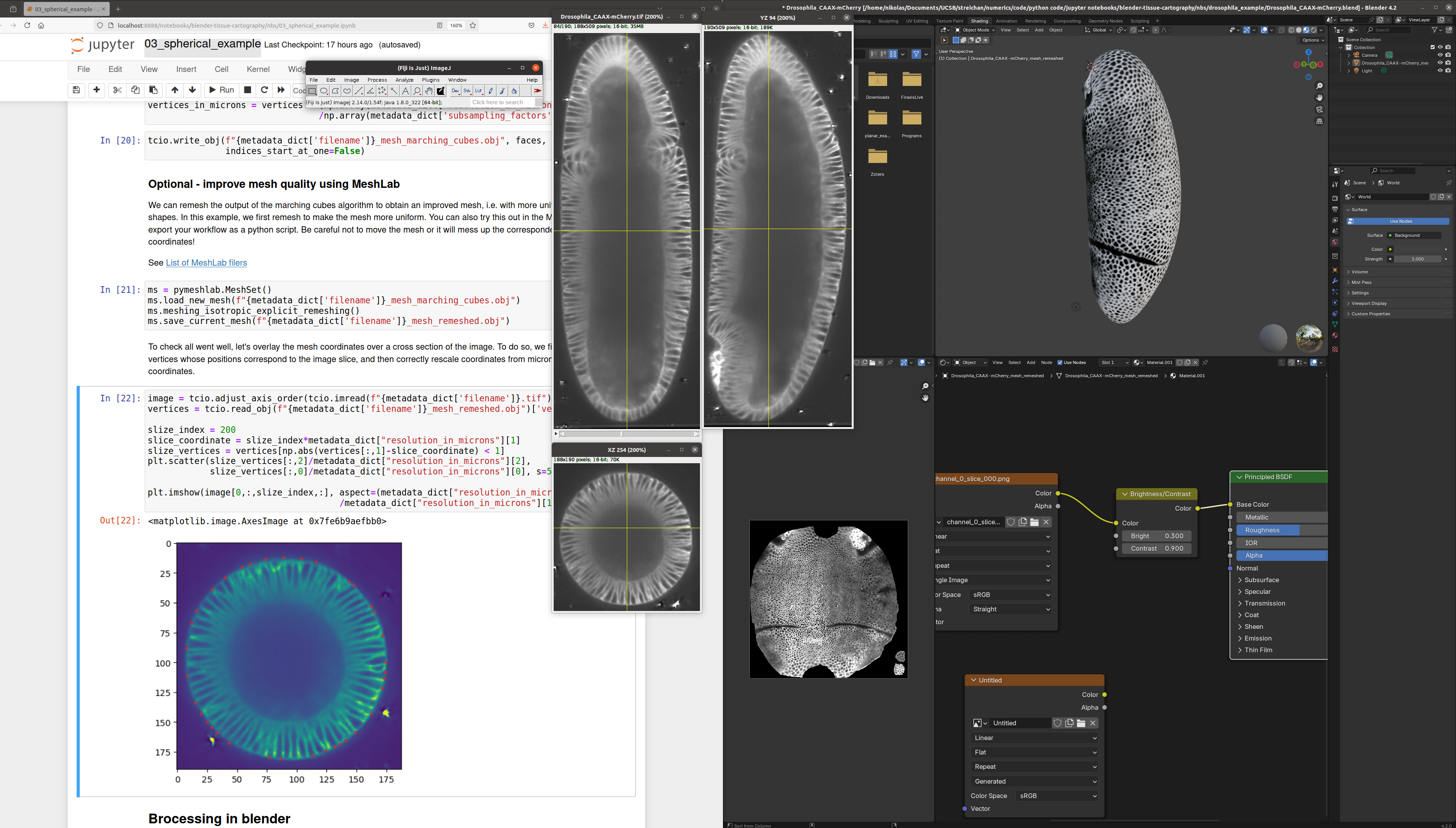
For advanced users, the blender_tissue_cartography library allows the creation of custom and automated tissue cartography pipelines, typically run from a jupyter computational notebook (which can also serve as lab notebook - notes, comments on the data). blender_tissue_cartography also provides tools for correct quantitative analysis of image data on curved surfaces.
Below is a screenshot to give you an idea of the workflow for the example Drosophila dataset: Volumetric data in ImageJ (center), jupyter computational notebook to run the blender_tissue_cartography module (left), and blender project with extracted mesh and texture (right).
Tutorials
Fully worked-out tutorials are provided on the documentation webpage. Test data for the tutorials can be downloaded from the nbs/Tutorials/ directory.
For the Python library, tutorials take the form of jupyter computational notebooks which you can download and run on your own computer (click the green button “Code” to download a .zip.) To run a tutorial on your computer, follow the installation instructions and then launch jupyter and work through the notebooks in the Tutorials directory in order. I recommended being comfortable with running simple Python code (you don’t have to do any coding yourself).
The tutorial notebooks can be used as templates for your analysis pipelines. Here is an example of a jupyter computational notebook (left), and the created projection visualized in Blender (right).
Notes for Python beginners
You will need a working Python installation (see here: installing anaconda/miniconda, and know how to launch jupyter notebooks. You will run the computational notebooks in your browser. Here is a video tutorial
Create a new folder for each tissue cartography project. Do not place them into the folder into which you unpacked
blender_tissue_cartography- otherwise, your files will be overwritten if you want to update the softwareThe repository contains two sets of notebooks: the
nbsfolder and thenbs/Tutorialsfolder. Thenbs-notebooks are for developing the code. If you don’t want to develop/adapt the code to your needs, you don’t need to look at them. Copy a notebook from thenbs/Tutorialsfolder - e.g.03_basics_example.ipynb- into your project folder to use it as a template.You do not need to copy functions into your notebooks manually. If you follow the installation instructions, the code will be installed as a Python package and can be “imported” by Python. See tutorials!
Software stack
Note: the Python libraries will be installed automatically if you follow the installation instructions above.
Required
Optional
Other useful software
- MicroscopyNodes plug-in for rendering volumetric
.tiffiles in blender - Boundary First Flattening advanced tool for creating UV maps with graphical and command line interface
- pyFM python library for mesh-to-mesh registration (for dynamic data) which may complement the algorithms that ship with
blender_tissue-cartography
Acknowledgements
This software is being developed by Nikolas Claussen in the Streichan lab at UCSB. We thank Cecile Regis, Susan Wopat, Noah Mitchell, An Yan, Pieter Derksen, Matthew Lefebvre, Sean Komura, Gary Han, Boris Fosso, and Dillon Cislo, for sharing data, advice, and software testing.
Citation
If you use blender_tissue_cartography for your work, please cite:
Nikolas Claussen, Cécile Regis, Susan Wopat, and Sebastian Streichan: Blender tissue cartography: an intuitive tool for the analysis of dynamic 3D microscopy data. bioRxiv 2025.02.04.636523; doi: https://doi.org/10.1101/2025.02.04.636523